Abstract
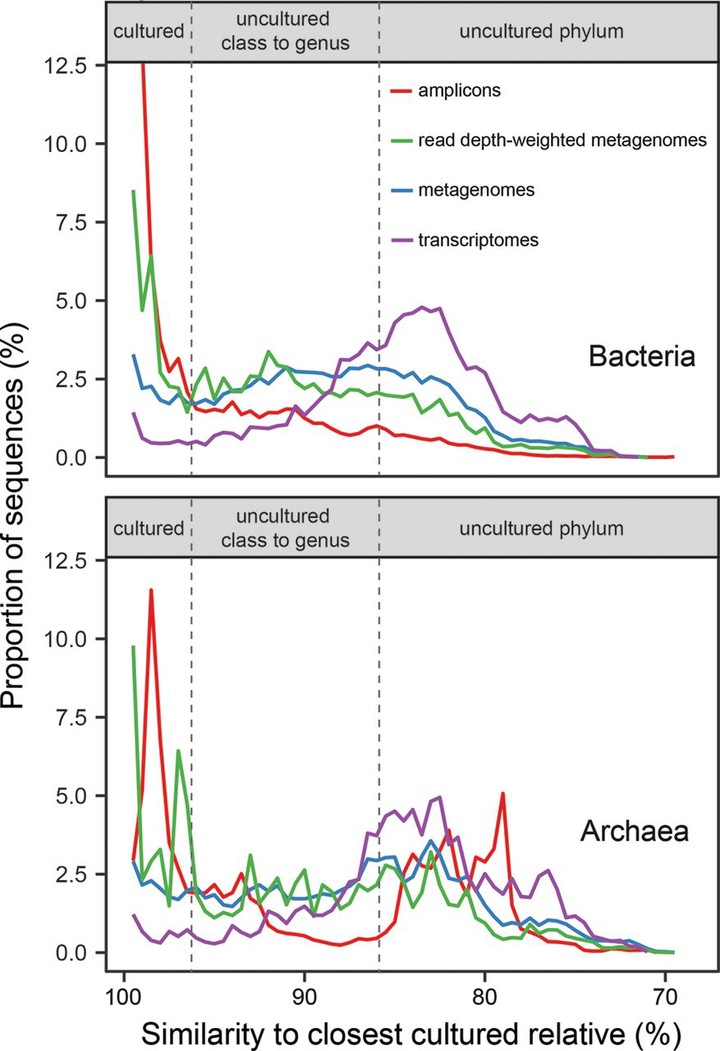
To describe a microbe’s physiology, including its metabolism, environmental roles, and growth characteristics, it must be grown in a laboratory culture. Unfortunately, many phylogenetically novel groups have never been cultured, so their physiologies have only been inferred from genomics and environmental characteristics. Although the diversity, or number of different taxonomic groups, of uncultured clades has been studied well, their global abundances, or numbers of cells in any given environment, have not been assessed. We quantified the degree of similarity of 16S rRNA gene sequences from diverse environments in publicly available metagenome and metatranscriptome databases, which we show have far less of the culture bias present in primer-amplified 16S rRNA gene surveys, to those of their nearest cultured relatives. Whether normalized to scaffold read depths or not, the highest abundances of metagenomic 16S rRNA gene sequences belong to phylogenetically novel uncultured groups in seawater, freshwater, terrestrial subsurface, soil, hypersaline environments, marine sediment, hot springs, hydrothermal vents, nonhuman hosts, snow, and bioreactors (22% to 87% uncultured genera to classes and 0% to 64% uncultured phyla). The exceptions were human and human-associated environments, which were dominated by cultured genera (45% to 97%). We estimate that uncultured genera and phyla could comprise 7.3 × 1029 (81%) and 2.2 × 1029 (25%) of microbial cells, respectively. Uncultured phyla were overrepresented in metatranscriptomes relative to metagenomes (46% to 84% of sequences in a given environment), suggesting that they are viable. Therefore, uncultured microbes, often from deeply phylogenetically divergent groups, dominate nonhuman environments on Earth, and their undiscovered physiologies may matter for Earth systems.