DNA and RNA sequence data are being deposited in public repositories at an astonishing rate. The quantity of sequence data in GenBank, for instance, has doubled approximately every 18 months since 1982! This is an extraordinarily rich source of information about microbial life. I argue that it is impossible to use all of the information in a sequence data set in a single study, and further that we can learn a lot by combining data from disparate studies that were not initially intended to be compared.
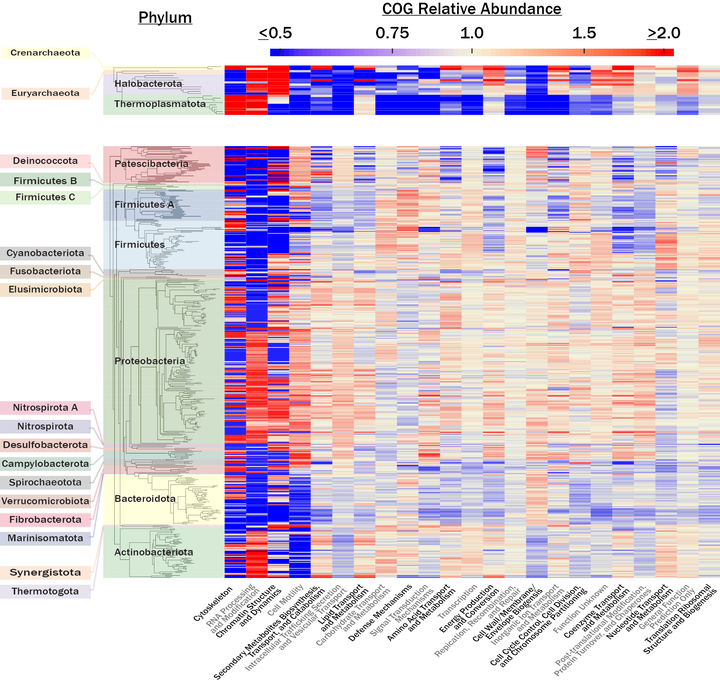
One line of research in the Steen Lab has been to see what we can learn from public data sets. We have helped to quantify the number of uncultured cells on Earth, the degree to which microbial traits are conserved as a function of taxonomic rank, and the relationship between DNA sequencing effort and the quantity of metagenome-assembled genomes (MAGs) that can be obtained.